

DNA marker 200-1500+ col.
Estimation of the size of DNA fragments generated in PCR (PCR fragments) or by DNA cutting by restriction enzymes (restriction fragments) is usually based on the comparison of these fragments with DNA fragments of known size (DNA markers). Size of PCR fragments is usually in the range 200-1500 base pairs. This range is covered by DNA fragments, which are part of DNA marker 200-1500+ coloured (Fig. 1). These fragments were produced by amplification of plasmid DNA of different length.
| Cat.no. | Name | Package | Price | |
|---|---|---|---|---|
| C130 | DNA marker 200-1500+ col. | 50 µg/300 µl | 1 590,00 Kč | |
| C131 | DNA marker 200-1500+ col. | 5x 50 µg/300 µl | 5 990,00 Kč | |
| C132 | DNA marker 200-1500+ col. | 20x 50 µg/300 µl | 19 990,00 Kč |
Product description
Estimation of the size of DNA fragments generated in PCR (PCR fragments) or by DNA cutting by restriction enzymes (restriction fragments) is usually based on the comparison of these fragments with DNA fragments of known size (DNA markers). Size of PCR fragments is usually in the range 200-1500 base pairs. This range is covered by DNA fragments, which are part of DNA marker 200-1500+ coloured (Fig. 1). These fragments were produced by amplification of plasmid DNA of different length.
Technical data
Concentration
- 1.7 µg DNA/10 µl buffer (10 mMTris-HCl,25 mMEDTA, additives).
Packaging
- 1 tube containing 50 µg of amplified DNA fragments in 300 µl of buffer. This amount enables preparation of 30 (each 10 µl) or 60 (each 5 µl) markers.
Storage
- Store at temperature -20 ± 5oC. Material can be repeatedly defrosted.
Quality control
- The presence of corresponding fragments is controlled by electrophoresis in agarose gel supplemented with ethidium bromide (1 µg/ml). When observed under UV light, 16 DNA fragments are observed (200, 250, 300, 350, 400, 450, 500, 550, 600, 650, 700, 800, 900, 1000, 1200, 1500 bp). 500 bp fragment is present in 2x higher amount (Fig. 1).
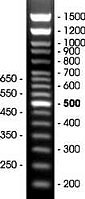
Figure. 1. Electrophoretic separation of DNA marker 200-1500+ coloured.
10 µl of DNA marker was directly loaded into wells of 1.2 agarose gel, which contained ethidium bromid (1 µg/ml) and 1x TBE buffer. DNA fragments were separated by electrophoresis. The numbers represent the number of base pairs in the corresponding DNA fragments.